Swagger Usage
Jump to navigation
Jump to search
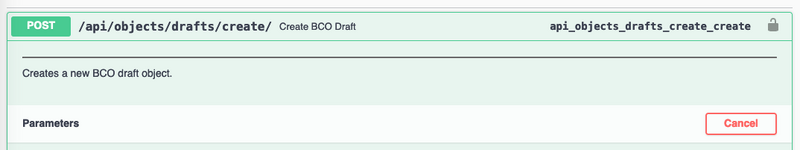
Submitting a BCO via Swagger
- Navigate to https://biocomputeobject.org/api/docs/#/BCO%20Management/api_objects_drafts_create_create
- Click on the lock in the top right of the picture below:
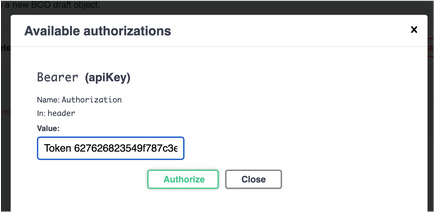
- Fill in the authorization box with your token as pictured below [NOTE! You *MUST* type ‘Token’ in the box as well as putting in your token] and click “authorize”.
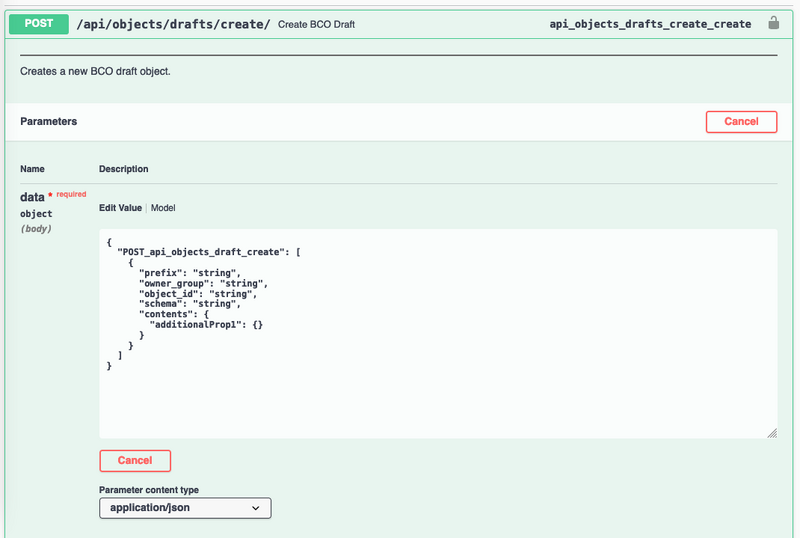
- Click on Edit Value
- Fill in the script:
- Prefix: Required, BCO Prefix to use
- Owner_group: Required, Group which owns the object and has permissions for it
- Schema: Required, This should be ‘IEEE’
- Contents: Required, BCO contents
- Example:
"POST_api_objects_draft_create": [
{
"prefix": "BCO",
"owner_group": "bco_drafter",
"schema": "IEEE",
"contents": {"etag":"06DACE70679F35BA87A3DD6FFFED4ED24A4F5B8C2571264C37E5F1B3ADE04A31","spec_version":"https://w3id.org/ieee/ieee-2791-schema/2791object.json","provenance_domain":{"name":"TESTING HCV1a ledipasvir resistance SNP detection","version":"2.9","review":[{"status":"approved","reviewer_comment":"Approved by GW staff. Waiting for approval from FDA Reviewer","date":"2017-11-12T12:30:48-0400","reviewer":{"name":"Charles Hadley King","affiliation":"George Washington University","email":"hadley_king@gwu.edu","contribution":["curatedBy"],"orcid":"https://orcid.org/0000-0003-1409-4549"}},{"status":"approved","reviewer_comment":"The revised BCO looks fine","date":"2017-12-12T12:30:48-0400","reviewer":{"name":"Eric Donaldson","affiliation":"FDA","email":"Eric.Donaldson@fda.hhs.gov","contribution":["curatedBy"]}}],"obsolete_after":"2118-09-26T14:43:43-0400","embargo":{"start_time":"2000-09-26T14:43:43-0400","end_time":"2000-09-26T14:43:45-0400"},"created":"2017-01-24T09:40:17-0500","modified":"2018-09-21T14:06:14-0400","contributors":[{"name":"Charles Hadley King","affiliation":"George Washington University","email":"hadley_king@gwu.edu","contribution":["createdBy","curatedBy"],"orcid":"https://orcid.org/0000-0003-1409-4549"},{"name":"Eric Donaldson","affiliation":"FDA","email":"Eric.Donaldson@fda.hhs.gov","contribution":["authoredBy"]}],"license":"https://spdx.org/licenses/CC-BY-4.0.html"},"usability_domain":["Identify baseline single nucleotide polymorphisms (SNPs)[SO:0000694], (insertions)[SO:0000667], and (deletions)[SO:0000045] that correlate with reduced (ledipasvir)[pubchem.compound:67505836] antiviral drug efficacy in (Hepatitis C virus subtype 1)[taxonomy:31646]","Identify treatment emergent amino acid (substitutions)[SO:1000002] that correlate with antiviral drug treatment failure","Determine whether the treatment emergent amino acid (substitutions)[SO:1000002] identified correlate with treatment failure involving other drugs against the same virus","GitHub CWL example: https://github.com/mr-c/hive-cwl-examples/blob/master/workflow/hive-viral-mutation-detection.cwl#L20"],"extension_domain":[{"fhir_extension":[{"fhir_endpoint":"http://fhirtest.uhn.ca/baseDstu3","fhir_version":"3","fhir_resources":[{"fhir_resource":"Sequence","fhir_id":"21376"},{"fhir_resource":"DiagnosticReport","fhir_id":"6288583"},{"fhir_resource":"ProcedureRequest","fhir_id":"25544"},{"fhir_resource":"Observation","fhir_id":"92440"},{"fhir_resource":"FamilyMemberHistory","fhir_id":"4588936"}]}]},{"scm_extension":{"scm_repository":"https://github.com/example/repo1","scm_type":"git","scm_commit":"c9ffea0b60fa3bcf8e138af7c99ca141a6b8fb21","scm_path":"workflow/hive-viral-mutation-detection.cwl","scm_preview":"https://github.com/example/repo1/blob/c9ffea0b60fa3bcf8e138af7c99ca141a6b8fb21/workflow/hive-viral-mutation-detection.cwl"}}],"description_domain":{"keywords":["HCV1a","Ledipasvir","antiviral resistance","SNP","amino acid substitutions"],"xref":[{"namespace":"pubchem.compound","name":"PubChem-compound","ids":["67505836"],"access_time":"2018-13-02T10:15-05:00"},{"namespace":"pubmed","name":"PubMed","ids":["26508693"],"access_time":"2018-13-02T10:15-05:00"},{"namespace":"so","name":"Sequence Ontology","ids":["SO:000002","SO:0000694","SO:0000667","SO:0000045"],"access_time":"2018-13-02T10:15-05:00"},{"namespace":"taxonomy","name":"Taxonomy","ids":["31646"],"access_time":"2018-13-02T10:15-05:00"}],"platform":["HIVE"],"pipeline_steps":[{"step_number":1,"name":"HIVE-hexagon","description":"Alignment of reads to a set of references","version":"1.3","prerequisite":[{"name":"Hepatitis C virus genotype 1","uri":{"uri":"http://www.ncbi.nlm.nih.gov/nuccore/22129792","access_time":"2017-01-24T09:40:17-0500"}},{"name":"Hepatitis C virus type 1b complete genome","uri":{"uri":"http://www.ncbi.nlm.nih.gov/nuccore/5420376","access_time":"2017-01-24T09:40:17-0500"}},{"name":"Hepatitis C virus (isolate JFH-1) genomic RNA","uri":{"uri":"http://www.ncbi.nlm.nih.gov/nuccore/13122261","access_time":"2017-01-24T09:40:17-0500"}},{"name":"Hepatitis C virus clone J8CF, complete genome","uri":{"uri":"http://www.ncbi.nlm.nih.gov/nuccore/386646758","access_time":"2017-01-24T09:40:17-0500"}},{"name":"Hepatitis C virus S52 polyprotein gene","uri":{"uri":"http://www.ncbi.nlm.nih.gov/nuccore/295311559","access_time":"2017-01-24T09:40:17-0500"}}],"input_list":[{"uri":"http://example.com/dna.cgi?cmd=objFile&ids=514683","access_time":"2017-01-24T09:40:17-0500"},{"uri":"http://example.com/dna.cgi?cmd=objFile&ids=514682","access_time":"2017-01-24T09:40:17-0500"}],"output_list":[{"uri":"http://example.com/data/514769/allCount-aligned.csv","access_time":"2017-01-24T09:40:17-0500"}]},{"step_number":2,"name":"HIVE-heptagon","description":"variant calling","version":"1.3","input_list":[{"uri":"http://example.com/data/514769/dnaAccessionBased.csv","access_time":"2017-01-24T09:40:17-0500"}],"output_list":[{"uri":"http://example.com/data/514801/SNPProfile.csv","access_time":"2017-01-24T09:40:17-0500"},{"uri":"http://example.com/data/14769/allCount-aligned.csv","access_time":"2017-01-24T09:40:17-0500"}]}]},"execution_domain":{"script":[{"uri":{"uri":"https://example.com/workflows/antiviral_resistance_detection_hive.py"}}],"script_driver":"shell","software_prerequisites":[{"name":"HIVE-hexagon","version":"babajanian.1","uri":{"uri":"http://example.com/dna.cgi?cmd=dna-hexagon&cmdMode=-","access_time":"2017-01-24T09:40:17-0500","sha1_checksum":"d60f506cddac09e9e816531e7905ca1ca6641e3c"}},{"name":"HIVE-heptagon","version":"albinoni.2","uri":{"uri":"http://example.com/dna.cgi?cmd=dna-heptagon&cmdMode=-","access_time":"2017-01-24T09:40:17-0500"}}],"external_data_endpoints":[{"name":"HIVE","url":"http://example.com/dna.cgi?cmd=login"},{"name":"access to e-utils","url":"http://eutils.ncbi.nlm.nih.gov/entrez/eutils/"}],"environment_variables":{"HOSTTYPE":"x86_64-linux","EDITOR":"vim"}},"parametric_domain":[{"param":"seed","value":"14","step":"1"},{"param":"minimum_match_len","value":"66","step":"1"},{"param":"divergence_threshold_percent","value":"0.30","step":"1"},{"param":"minimum_coverage","value":"15","step":"2"},{"param":"freq_cutoff","value":"0.10","step":"2"}],"io_domain":{"input_subdomain":[{"uri":{"filename":"Hepatitis C virus genotype 1","uri":"http://www.ncbi.nlm.nih.gov/nuccore/22129792","access_time":"2017-01-24T09:40:17-0500"}},{"uri":{"filename":"Hepatitis C virus type 1b complete genome","uri":"http://www.ncbi.nlm.nih.gov/nuccore/5420376","access_time":"2017-01-24T09:40:17-0500"}},{"uri":{"filename":"Hepatitis C virus (isolate JFH-1) genomic RNA","uri":"http://www.ncbi.nlm.nih.gov/nuccore/13122261","access_time":"2017-01-24T09:40:17-0500"}},{"uri":{"uri":"http://www.ncbi.nlm.nih.gov/nuccore/386646758","access_time":"2017-01-24T09:40:17-0500"}},{"uri":{"filename":"Hepatitis C virus S52 polyprotein gene","uri":"http://www.ncbi.nlm.nih.gov/nuccore/295311559","access_time":"2017-01-24T09:40:17-0500"}},{"uri":{"filename":"HCV1a_drug_resistant_sample0001-01","uri":"http://example.com/nuc-read/514682","access_time":"2017-01-24T09:40:17-0500"}},{"uri":{"filename":"HCV1a_drug_resistant_sample0001-02","uri":"http://example.com/nuc-read/514683","access_time":"2017-01-24T09:40:17-0500"}}],"output_subdomain":[{"mediatype":"text/csv","uri":{"uri":"http://example.com/data/514769/dnaAccessionBased.csv","access_time":"2017-01-24T09:40:17-0500"}},{"mediatype":"text/csv","uri":{"uri":"http://example.com/data/514801/SNPProfile*.csv","access_time":"2017-01-24T09:40:17-0500"}}]},"error_domain":{"empirical_error":{"false_negative_alignment_hits":"<0.0010","false_discovery":"<0.05"},"algorithmic_error":{"false_positive_mutation_calls_discovery":"<0.00005","false_discovery":"0.005"}}}
}
]
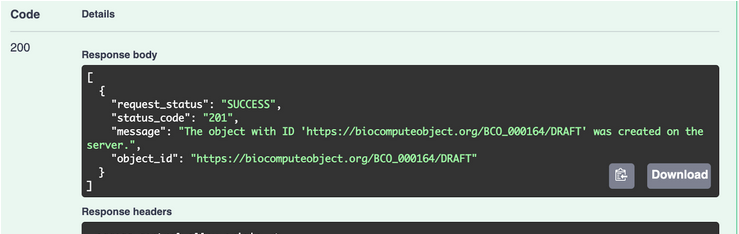
- Hit the EXECUTE button to submit the request.
- If successfully, should look like this:
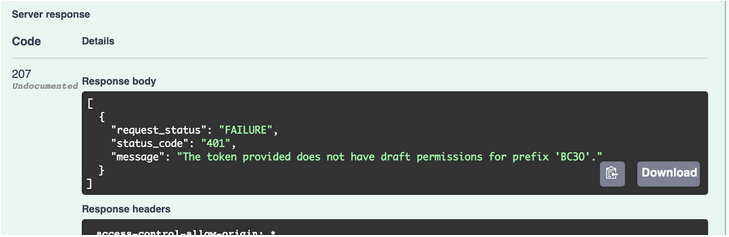
- If unsuccessful, will look like this: