Galaxy workflow converter: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
(→Upload) |
||
| Line 16: | Line 16: | ||
== Upload == | == Upload == | ||
# Converted JSON file can be uploaded to BCODB through copy/pasting the RAW JSON file directly on BCO Builder or through API, refer to API documentation [[BCO Portal Local Deployment|here]] | # Converted JSON file can be uploaded to BCODB through copy/pasting the RAW JSON file directly on BCO Builder or through API, refer to API documentation [[BCO Portal Local Deployment|here]]. | ||
Revision as of 21:00, 10 November 2022
Workflows can be directly downloaded from Galaxy but file conversion is needed in order to transfer into BCODB successfully. Please follow the following steps to convert the file and validation:
File conversion
- Download the converter script from GitHub.
- On the local terminal, run the following command
python [converter script's name.py] -b [path to the file that needs to be converted] -o [path to the output file]
Validation
- Install Validation Package:
- On the local terminal, clone the repository first by
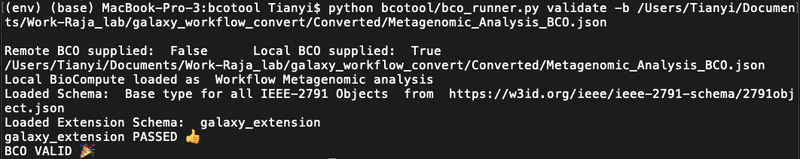
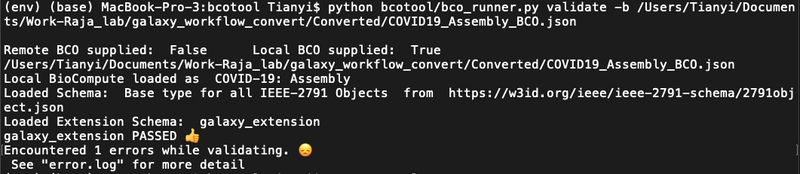
git clone https://github.com/biocompute-objects/bcotool cd bcotool/python -m venv env. env/bin/activatepip install -r requirements.txtpython bcotool/bco_runner.py validate -b [path to the converted file](Note: if only runpython bcotool/bco_runner.py, more options can be seen.) If passed validation should show as the figure below- If they encountered errors, will show as the figure below:
- Open error log for error description:
open error.log
- Open error log for error description:
- On the local terminal, clone the repository first by
Upload
- Converted JSON file can be uploaded to BCODB through copy/pasting the RAW JSON file directly on BCO Builder or through API, refer to API documentation here.