BCO Portal Local Deployment: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
No edit summary |
||
| Line 41: | Line 41: | ||
#Run server: <code>python3.9 manage.py runserver 8181</code> | #Run server: <code>python3.9 manage.py runserver 8181</code> | ||
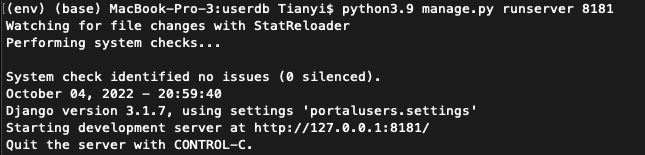
##If ran successfully, you should observe as shown in the following figure:[[File:Screen Shot 2022-10-05 at 14.20.20.png|800px|thumb|center]] | ##If ran successfully, you should observe as shown in the following figure:[[File:Screen Shot 2022-10-05 at 14.20.20.png|800px|thumb|center]] | ||
##Once see the messages as shown in the figure above, go to http://localhost:8181/users/admin/ on the local browser to see if BCO API is set up successfully | ##Once see the messages as shown in the figure above, go to http://localhost:8181/users/admin/ on the local browser to see if BCO API is set up successfully (Note: If it worked you should be able to login using the SuperUser credentials you created above) | ||
Revision as of 18:56, 5 October 2022
In order to work with BCO API locally on a Mac OS, please follow the following steps to set up:
BCO_API:
- Clone the GitHub Repository
- Open Terminal, enter the following commands:
git clone https://github.com/biocompute-objects/bco_apicd bco_api
- Open Terminal, enter the following commands:
- Switch to the latest release branch:
git switch 22.10
- Enter the repository, create a virtual environment, and install the required packages:
pyenv local 3.9.4(Note: will show as pyenv: command not found if already installed)python3.9 -m venv envsource env/bin/activate(activated virtual environment by showing (env) (base) MacBook-Pro-3:bco_api)python -m pip install -r requirements.txt(if installation failed, make sure you are in the correct path: /Users/Username/bco_api)
- Modify Configuration file
- Once in the virtual environment, make sure you are on the path:/Users/Username/bco_api
- Edit the server.conf file:
vim bco_api/bco_api/server.conf
- Quick check to make sure the server can run
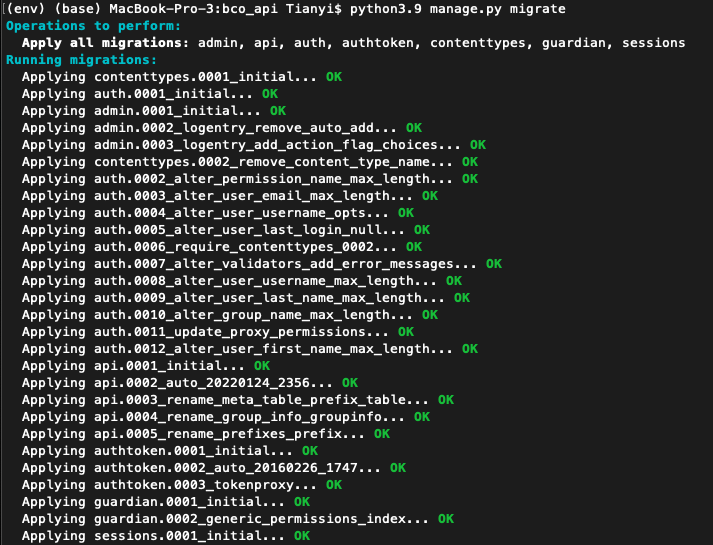
cd bco_api/bco_api(Note after this command, should under path /Users/Username/bco_api/bco_api)- Make migrations:
python3.9 manage.py migrate - If run successfully, should observe the following:
- Create a super user for the API:
python3.9 manage.py createsuperuser- Follow the prompts
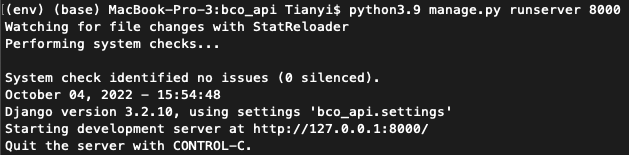
python3.9 manage.py runserver 8000:- Once see the messages as shown in the figure above, go to http://localhost:8000/api/admin/ on the local browser to see if BCO API is set up successfully
UserDB
- Clone the GitHub Repository
- Open Terminal, enter the following commands:
git clone https://github.com/biocompute-objects/userdbcd userdb/
- Open Terminal, enter the following commands:
- Switch to the desired branch: e.g.
git switch 22.10 - Enter the repository, create a virtual environment, and install the required packages:
pyenv local 3.9.4python3.9 -m venv envsource env/bin/activatepython -m pip install -r requirements.txt
- Modify the Config file:
vim portalusers/settings.py - Make migrations:
python3.9 manage.py migrate - Create a super user for the API:
python3.9 manage.py createsuperuser(Note: this step can be skipped if superuser is already created or not superuser account is not necessary) - Run server:
python3.9 manage.py runserver 8181- If ran successfully, you should observe as shown in the following figure:
- Once see the messages as shown in the figure above, go to http://localhost:8181/users/admin/ on the local browser to see if BCO API is set up successfully (Note: If it worked you should be able to login using the SuperUser credentials you created above)