BCO Portal Local Deployment: Difference between revisions
Jump to navigation
Jump to search
| Line 76: | Line 76: | ||
## <code>source env/bin/activate</code> | ## <code>source env/bin/activate</code> | ||
## <code>pip3.9 install -r requirements.txt</code> | ## <code>pip3.9 install -r requirements.txt</code> | ||
# Check the Configuration files: | |||
## Edit the server.conf file: <code>vim /var/www/bcoeditor/bco_api/bco_api/server.conf</code> | |||
### Change the version number to match the version you are modifying | |||
# Make sure the server can run: <code>cd /var/www/bcoeditor/bco_api/bco_api</code> | |||
# Make Migrations: | |||
## python3.9 manage.py migrate | |||
## python3.9 manage.py collectstatic | |||
## python3.9 manage.py runserver 8000 | |||
## Go to https://test.portal.biochemistry.gwu.edu/api/admin/ to make changes | |||
## Ctrl+C to exit the running server | |||
Revision as of 19:30, 27 October 2022
Configuration
In order to work with BCO API locally on a Mac OS, please follow the following steps to set up:
BCO_API
- Clone the GitHub Repository
- Open Terminal, enter the following commands:
git clone https://github.com/biocompute-objects/bco_apicd bco_api
- Open Terminal, enter the following commands:
- Switch to the latest release branch:
git switch 22.10
- Enter the repository, create a virtual environment, and install the required packages:
pyenv local 3.9.4(Note: will show as pyenv: command not found if already installed)python3.9 -m venv envsource env/bin/activate(activated virtual environment by showing (env) (base) MacBook-Pro-3:bco_api)python -m pip install -r requirements.txt(if installation failed, make sure you are in the correct path: /Users/Username/bco_api)
- Modify Configuration file
- Once in the virtual environment, make sure you are on the path:/Users/Username/bco_api
- Edit the server.conf file:
vim bco_api/bco_api/server.conf
- Quick check to make sure the server can run
cd bco_api/bco_api(Note after this command, should under path /Users/Username/bco_api/bco_api)- Make migrations:
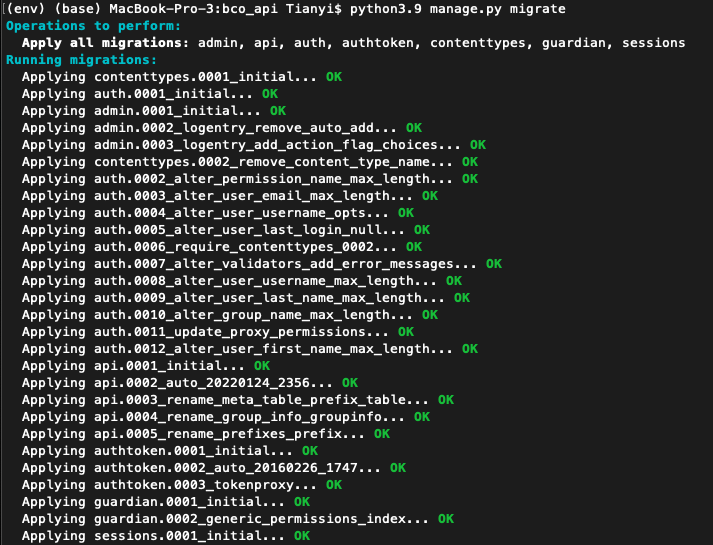
python3.9 manage.py migrate - If run successfully, should observe the following:
- Create a super user for the API:
python3.9 manage.py createsuperuser- Follow the prompts
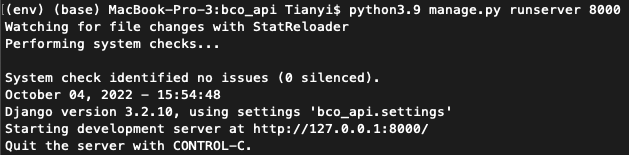
python3.9 manage.py runserver 8000:- Once see the messages as shown in the figure above, go to http://localhost:8000/api/admin/ on the local browser to see if BCO API is set up successfully
UserDB
- Clone the GitHub Repository
- Open Terminal, enter the following commands:
git clone https://github.com/biocompute-objects/userdbcd userdb/
- Open Terminal, enter the following commands:
- Switch to the desired branch: e.g.
git switch 22.10 - Enter the repository, create a virtual environment, and install the required packages:
pyenv local 3.9.4python3.9 -m venv envsource env/bin/activatepython -m pip install -r requirements.txt
- Modify the Config file:
vim portalusers/settings.py - Make migrations:
python3.9 manage.py migrate - Create a super user for the API:
python3.9 manage.py createsuperuser(Note: this step can be skipped if a superuser is already created or not superuser account is not necessary) - Run server:
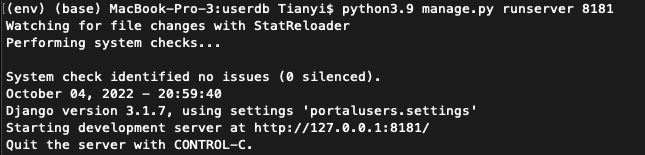
python3.9 manage.py runserver 8181- If ran successfully, you should observe as shown in the following figure:
- Once see the messages as shown in the figure above, go to http://localhost:8181/users/admin/ on the local browser to see if BCO API is set up successfully (Note: If it worked you should be able to login using the SuperUser credentials you created above)
Portal Deployment Instructions
- Clone the repository
- git clone https://github.com/biocompute-objects/portal
- cd portal/
- Switch to the desired branch: e.g.
git switch 22.10 - Install packages with either of the following commands:
npm installnpm install --legacy-peer-deps
- Edit /portal/src/App.js line 87 based on your need :
- For local,
sending = hostnames.local - For the test site,
sending = hostnames.test - For the production site,
sending = hostnames.production
- For local,
- Start running:
npm run start
Production Portal Deployment
The BCO API repository contains a top-level folder “admin_only” which contains service definitions for gunicorn and Django. Thus, we can first clone the repository, then perform a few administrative steps to start the API.
- Connect to 'go.vpn.gwu.edu/SMHS_BIOC' VPN first.
- Enter test/production server via:
ssh USERNAME@test.portal.biochemistry.gwu.edu(for test server)/ssh USERNAME@portal.biochemistry.gwu.edu(for production server) (Note: /home and /etc are shared between the two servers) - Backup and archive DB:
cd /var/www/bcoeditor/db_backupscp ../bco_api/bco_api/db.sqlite3 db.sqlite3.bak.api.[TODAY'S DATE] (e.g. cp ../bco_api/bco_api/db.sqlite3 db.sqlite3.bak.api.22.10.25)cp ../userdb/db.sqlite3 db.sqlite3.bak.udb.[TODAY'S DATE]cp ../bco_api/bco_api/server.conf server.conf.bak.[TODAY'S DATE]
- Pull the repository
cd /var/www/bcoeditor/bco_apigit clone https://github.com/biocompute-objects/bco_api(if cloned already, step not needed)
- Enter the repository, create a virtual environment and install the required packages:
- First switch to the proper user account via
su - bco_api_user(If need to use this account, please contact BCO team for password) - Switch to the desired branch:
git switch 22.11 source env/bin/activatepip3.9 install -r requirements.txt
- First switch to the proper user account via
- Check the Configuration files:
- Edit the server.conf file:
vim /var/www/bcoeditor/bco_api/bco_api/server.conf- Change the version number to match the version you are modifying
- Edit the server.conf file:
- Make sure the server can run:
cd /var/www/bcoeditor/bco_api/bco_api - Make Migrations:
- python3.9 manage.py migrate
- python3.9 manage.py collectstatic
- python3.9 manage.py runserver 8000
- Go to https://test.portal.biochemistry.gwu.edu/api/admin/ to make changes
- Ctrl+C to exit the running server