BCO Pilot Project: Difference between revisions
ChinweokeO (talk | contribs) (Created page with "== Introduction == Overview: Briefly introduce the BioCompute Pilot Project, its purpose, and significance. Objectives: Outline the key goals and objectives of the project. Scope: Define the scope of the project, including the types of data and analyses involved. == BioCompute Objects (BCOs) == What is a BCO?: Provide a detailed explanation of BioCompute Objects, including their purpose and components. Structure of a BCO: Break down the sections of a BCO, such as Pro...") |
No edit summary |
||
| (19 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
== Pilot Project == | == Pilot Project == | ||
BioCompute is shorthand for IEEE 2791-2020, a [https://standards.ieee.org/ieee/2791/7337/ standard for communicating computational workflows]. The standard was developed in response to difficulties in recording and interpreting bioinformatics pipelines. BioCompute is a framework for step-by-step description of computational workflows. BioCompute defines what content must be included, and what form it should take. It includes data provenance and rich metadata to eliminate ambiguity. The standard also includes an “Execution Domain” for re-executing a pipeline, if desired. | |||
Three Centers at the FDA [https://www.federalregister.gov/documents/2020/07/22/2020-15771/electronic-submissions-data-standards-support-for-the-international-institute-of-electrical-and have adopted the standard] (see notice in Federal Registry by the Food and Drug Administration on 07/22/2020) for bioinformatics applications. To increase adoption of BioCompute and bring awareness within and outside of FDA we propose a pilot with the BioCompute team, FDA, and industry. Because the standard was developed to manage complexity and reduce the organizational burden on FDA reviewers and regulated industry submitting to them, the purpose of this pilot project is to bring both sides in agreement around its usage thereby streamlining and standardizing computational workflow submissions and reviews. Participating sponsors will prepare and submit a computational workflow with a BioCompute Object (BCO) without any additional clarification, and FDA reviewers will review the BCO. Initial planning meetings, gathering project materials, preparing and submitting BCOs, BCO review, and final debrief are expected to take six months to one year. The overall project will be coordinated by George Washington University (GW) BioCompute team. | |||
== Expected Outcome == | |||
=== Documentation and Reporting === | |||
Throughout the process, we will document anything difficult to understand, barriers to working with or reading the BCO, and any other general issues. As a result, a [https://wiki.biocomputeobject.org/Best_Practices best practices document] and [https://wiki.biocomputeobject.org/Security_Plan security plan] have been generated to support the standard and there may be additional training materials, created as needed to enable a group to prepare or read a BCO with minimal training. These resources are meant to support the standard as a vehicle for more fluid communication and to enable a group to prepare or read a BCO with minimal training. | |||
In addition, groups that participate in the pilot will have first-hand knowledge of submitting computational analyses and how to do so efficiently. Monthly reports are also submitted to maintain communication of plans and processes as they occur. | |||
== Request == | |||
=== Collaboration and Communication === | |||
To have the most impact, this project requires input from multiple groups with potential collaboration with the FDA-HIVE team in CBER. We’re asking FDA reviewers who review any work that contains a computational pipeline to participate. Examples of computational pipelines include a medical device for the detection of physiological warning signs, a diagnostic test for the presence of pathogenic species, supporting evidence for a new drug application in the form of NGS analyses, or others. | |||
Only the computational portion needs to be submitted, and participants will use their data and workflow (real or synthetic mock data and workflow) for submission. Data submitted need not be actual clinical data, however, the more similar the data to a real submission, the more valuable the pilot. Participating FDA reviewers will be asked to review the pilot submission with no additional input, and to document anything they feel is unclear or not presented well. | |||
Participating sponsors are asked to prepare realistic regulatory submissions using one or more BCOs to describe their work. We estimate that the project will require some level of effort from all participants. The representative(s) will be involved in planning, gathering the materials for submission, and learning how to represent their data in the BCO, as well as meetings and documentation for the pilot project. | |||
Groups are asked to meet with us to explain difficulties working with the standard. Groups may decide to meet with us individually, if they desire, rather than as part of a larger meeting with the other groups. | |||
=== Estimated Contributions === | |||
{| class="wikitable" | |||
!'''Organization''' | |||
'''Platform''' | |||
!'''Personnel''' | |||
!'''Tasks''' | |||
|- | |||
|Velsera BCO platform | |||
|Coordinator | |||
|Project Liaison with technical team, platform representative | |||
|- | |||
|DNAnexus BCO platform | |||
|Coordinator | |||
|Project Liaison with technical team, platform representative | |||
|- | |||
|FDA-HIVE BCO platform | |||
|Reviewer/Technologist | |||
|Review submissions, document feedback | |||
|- | |||
|GW BCOdb and BCO-form | |||
|Project Manager+Technical Lead for BCOdb and form-based submission) | |||
|Overall plan, organize regular meetings/agendas/schedules, project documentation | |||
|- | |||
|Industry Participants (3 total; will be paired with GW, SBG and DNAnexus) | |||
|Coordinator | |||
|Gather materials, coordinate pilot submission internally | |||
|- | |||
|FDA | |||
|Reviewer | |||
|Document feedback | |||
|} | |||
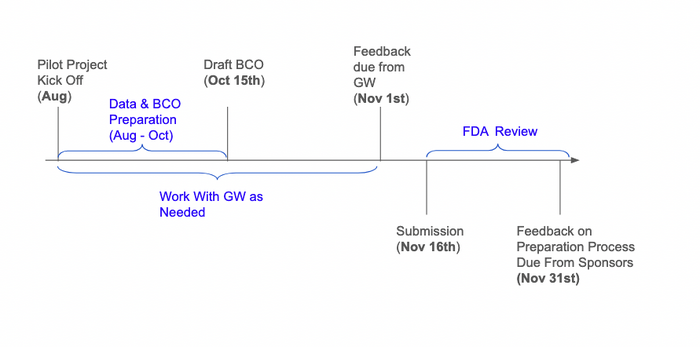
=== Estimated Timeline === | |||
[[File:Screenshot 2024-08-26 at 14.34.23.png|center|thumb|700x700px]] | |||
=== | |||
=== Challenges and Solutions === | === Challenges and Solutions === | ||
During development and maintenance of the platform, users may run into bugs and may have suggestions regarding the user interface (UI). To address these challenges, users can submit their feedback and report issues via the website's [https://github.com/biocompute-objects/portal_userdb/issues/new/choose 'Report an Issue'] link. This feedback is useful in refining the platform and enhancing the overall user experience. Additionally, to ensure continuous effectiveness and functionality of BioCompute Objects (BCOs), regular updates and enhancements are made based on testing and user feedback. | |||
== Frequently Asked Questions == | |||
For all submission related questions and general BCO usage, refer to the [[FAQs#Submitting BCOs to FDA|FAQ]] page. | |||
== References and Resources == | |||
For additional information, please refer to the resources below: | |||
= | * [https://wiki.biocomputeobject.org/index.php?title=Main_Page BioCompute Objects Wiki] | ||
* [https://wiki.biocomputeobject.org/White_paper White Paper] | |||
=== Contact === | |||
For any questions or to participate in the pilot, please email Jonathon Keeney, project co-PI at George Washington University: keeneyjg@gwu.edu | |||
Latest revision as of 19:14, 9 September 2024
Pilot Project
BioCompute is shorthand for IEEE 2791-2020, a standard for communicating computational workflows. The standard was developed in response to difficulties in recording and interpreting bioinformatics pipelines. BioCompute is a framework for step-by-step description of computational workflows. BioCompute defines what content must be included, and what form it should take. It includes data provenance and rich metadata to eliminate ambiguity. The standard also includes an “Execution Domain” for re-executing a pipeline, if desired.
Three Centers at the FDA have adopted the standard (see notice in Federal Registry by the Food and Drug Administration on 07/22/2020) for bioinformatics applications. To increase adoption of BioCompute and bring awareness within and outside of FDA we propose a pilot with the BioCompute team, FDA, and industry. Because the standard was developed to manage complexity and reduce the organizational burden on FDA reviewers and regulated industry submitting to them, the purpose of this pilot project is to bring both sides in agreement around its usage thereby streamlining and standardizing computational workflow submissions and reviews. Participating sponsors will prepare and submit a computational workflow with a BioCompute Object (BCO) without any additional clarification, and FDA reviewers will review the BCO. Initial planning meetings, gathering project materials, preparing and submitting BCOs, BCO review, and final debrief are expected to take six months to one year. The overall project will be coordinated by George Washington University (GW) BioCompute team.
Expected Outcome
Documentation and Reporting
Throughout the process, we will document anything difficult to understand, barriers to working with or reading the BCO, and any other general issues. As a result, a best practices document and security plan have been generated to support the standard and there may be additional training materials, created as needed to enable a group to prepare or read a BCO with minimal training. These resources are meant to support the standard as a vehicle for more fluid communication and to enable a group to prepare or read a BCO with minimal training.
In addition, groups that participate in the pilot will have first-hand knowledge of submitting computational analyses and how to do so efficiently. Monthly reports are also submitted to maintain communication of plans and processes as they occur.
Request
Collaboration and Communication
To have the most impact, this project requires input from multiple groups with potential collaboration with the FDA-HIVE team in CBER. We’re asking FDA reviewers who review any work that contains a computational pipeline to participate. Examples of computational pipelines include a medical device for the detection of physiological warning signs, a diagnostic test for the presence of pathogenic species, supporting evidence for a new drug application in the form of NGS analyses, or others.
Only the computational portion needs to be submitted, and participants will use their data and workflow (real or synthetic mock data and workflow) for submission. Data submitted need not be actual clinical data, however, the more similar the data to a real submission, the more valuable the pilot. Participating FDA reviewers will be asked to review the pilot submission with no additional input, and to document anything they feel is unclear or not presented well.
Participating sponsors are asked to prepare realistic regulatory submissions using one or more BCOs to describe their work. We estimate that the project will require some level of effort from all participants. The representative(s) will be involved in planning, gathering the materials for submission, and learning how to represent their data in the BCO, as well as meetings and documentation for the pilot project.
Groups are asked to meet with us to explain difficulties working with the standard. Groups may decide to meet with us individually, if they desire, rather than as part of a larger meeting with the other groups.
Estimated Contributions
| Organization
Platform |
Personnel | Tasks |
|---|---|---|
| Velsera BCO platform | Coordinator | Project Liaison with technical team, platform representative |
| DNAnexus BCO platform | Coordinator | Project Liaison with technical team, platform representative |
| FDA-HIVE BCO platform | Reviewer/Technologist | Review submissions, document feedback |
| GW BCOdb and BCO-form | Project Manager+Technical Lead for BCOdb and form-based submission) | Overall plan, organize regular meetings/agendas/schedules, project documentation |
| Industry Participants (3 total; will be paired with GW, SBG and DNAnexus) | Coordinator | Gather materials, coordinate pilot submission internally |
| FDA | Reviewer | Document feedback |
Estimated Timeline
Challenges and Solutions
During development and maintenance of the platform, users may run into bugs and may have suggestions regarding the user interface (UI). To address these challenges, users can submit their feedback and report issues via the website's 'Report an Issue' link. This feedback is useful in refining the platform and enhancing the overall user experience. Additionally, to ensure continuous effectiveness and functionality of BioCompute Objects (BCOs), regular updates and enhancements are made based on testing and user feedback.
Frequently Asked Questions
For all submission related questions and general BCO usage, refer to the FAQ page.
References and Resources
For additional information, please refer to the resources below:
Contact
For any questions or to participate in the pilot, please email Jonathon Keeney, project co-PI at George Washington University: keeneyjg@gwu.edu